Prevalence of non- representation of HLA antigens and alleles in Single Antigen Bead assay panels in patients at a referral lab
Shruti Tapiawala1,2, Suchita P Jogale2, Prashant Rajput1, Zaheer Virani1, Hepal Vora1, Bharat Shah1,2.
1Institute of Renal Sciences, Gleneagles Hospital, Mumbai, India; 2Histocompatibility, ATDI Lab Pvt Ltd, Mumbai, India
Introduction: Identification and monitoring of HLA antibodies against the Donor HLA antigens are one of the ways to risk stratify and monitor an allograft. The single antigen bead (SAB) assay serves as a extensive virtual cross-match tool to help a laboratory and a clinician treat a patient.
It is widely acknowledged that there is HLA allele variability across ethnicities and genetic backgrounds. This necessitates a thorough representation of prevalent alleles in the population on the beads used for the SAB assay. Deficiency of representation can lead to a false negative virtual crossmatch and may lead to immune events in recipients if the clinical teams are not aware of the presence of DSAs. We aimed to identify the prevalence of non- representation of HLA antigens and alleles in our population of patients
Material & Methods: 488 consecutive patients referred for HLA typing and SAB analysis in the period of August 2021 to February 2024 were studied. Their HLA antigens and alleles were analysed for their representation on the SAB panels. Intermediate resolution HLA typing of 6 Loci A,B,C, DRB1,2,3,4,5,DP and DQ by SSOP method performed on Luminex platform of these 488 individuals was compared with the HLA antigens and alleles available on the SAB panels from both the existing vendors A and B.
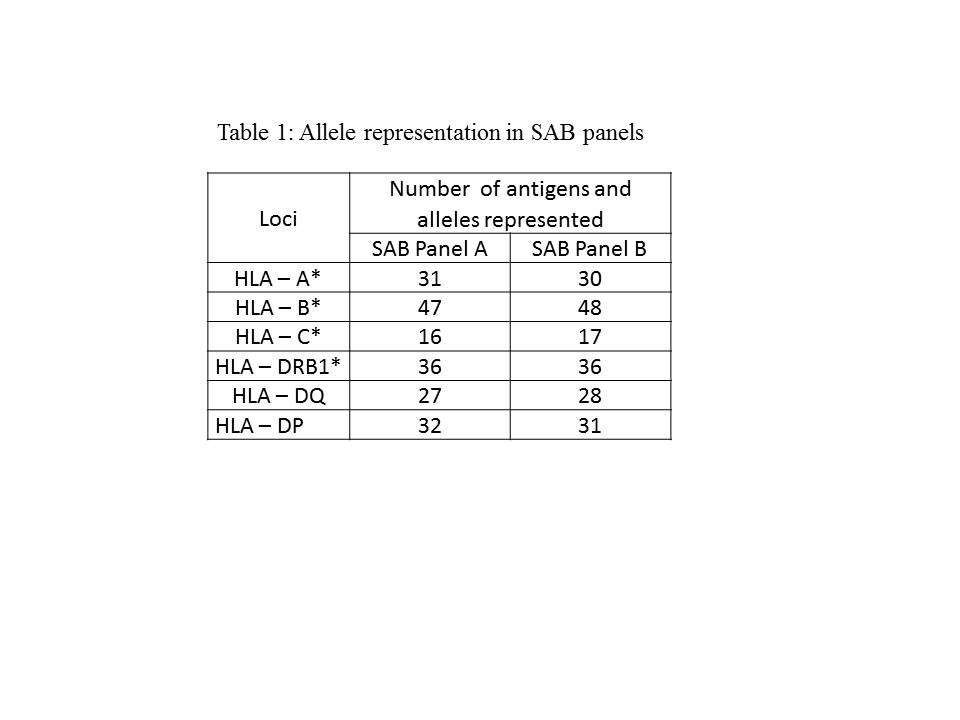
Results: A total of 185 distinct alleles were identified in the study population. Among these, 66.48% (n = 123) of alleles were represented in SAB panel A and 64.86% (n=120) in SAB panel B. The unrepresented alleles accounted for 33.51% (n = 62) in SAB panel A and 35.13% (n= 65) in SAB panel B. Whereas 25.4% (n= 47) alleles were not represented in both the SAB panels. Table 1 represents the comparative analysis of allele representation in SAB panels.
Conclusion: Our study highlights the limitation of relying on the presently available SAB panels to identify the presence of DSAs. One cannot rely on virtual crossmatch of the donor HLA with recipient HLA antibody profile performed with existing SAB panels due to the non-representation of locally prevalent or unique alleles in our population. Consequently, we advocate combining SAB profile with physical crossmatching when encountering mismatched alleles that lack representation, until a comprehensive allele coverage or robust epitope matching is achieved.
