Jin-Myung Kim, Korea has been granted the TTS-KST Basic and Translational Mentee-Mentor Award
Exploring the molecular pathways of intracranial aneurysm formation in autosomal dominant polycystic kidney disease using proteomic analysis
Jin-Myung Kim1, Hee-Sung Ahn2, Hye Eun Kwon1, Youngmin Ko1, Joo Hee Jung1, Hyunwook Kwon1, Young Hoon Kim1, Kyunggon Kim3, Sung Shin1.
1Department of Surgery, Asan Medical Center, Seoul, Korea; 2AMC sciences, Asan Medical Center, Seoul, Korea; 3Department of Convergence Medicine, Asan Medical Cen, Seoul, Korea
Introduction: Intracranial aneurysm (IA) frequently coincides with autosomal dominant polycystic kidney disease (ADPKD), exhibiting incidence rates nearly 10 times higher than the general population. However, the exact mechanism of how these two conditions are related remains unclear. This study aims to identify mechanisms behind IA occurrence in ADPKD patients using proteomics and to discover potential protein biomarkers for early diagnosis.
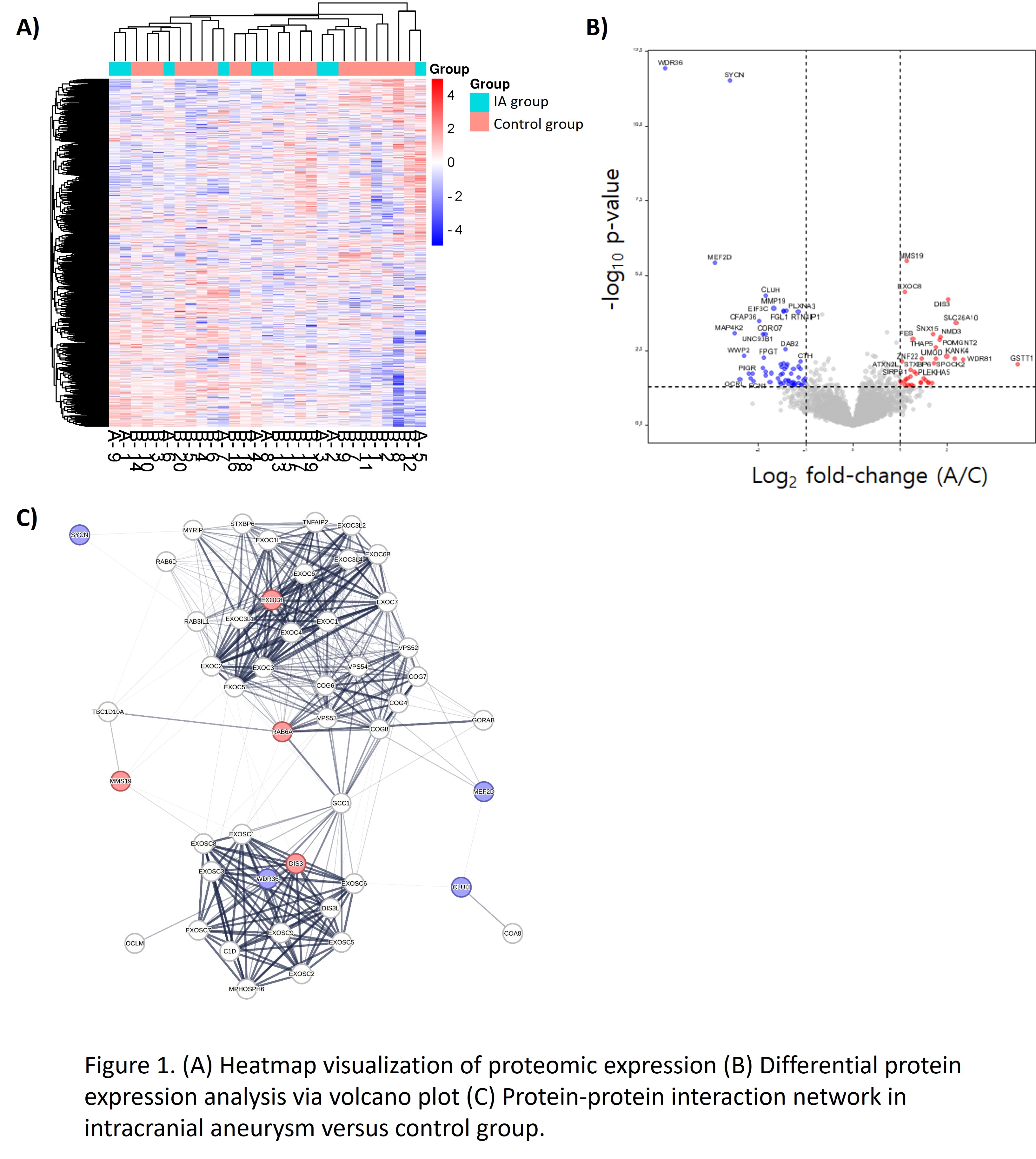
Method: Pre-kidney transplantation ADPKD patients underwent cranial CT and/or MR angiography, with findings dictating assignment to either a control group (ADPKD without IA, n=20), an IA group (ADPKD with IA, n=9). During transplantation, bilateral nephrectomy was performed and native renal arteries were sampled for proteomic analysis via a liquid chromatography-tandem mass spectrometry. Differentially expressed proteins were subjected to bioinformatic analysis and a protein-protein interaction network analysis.
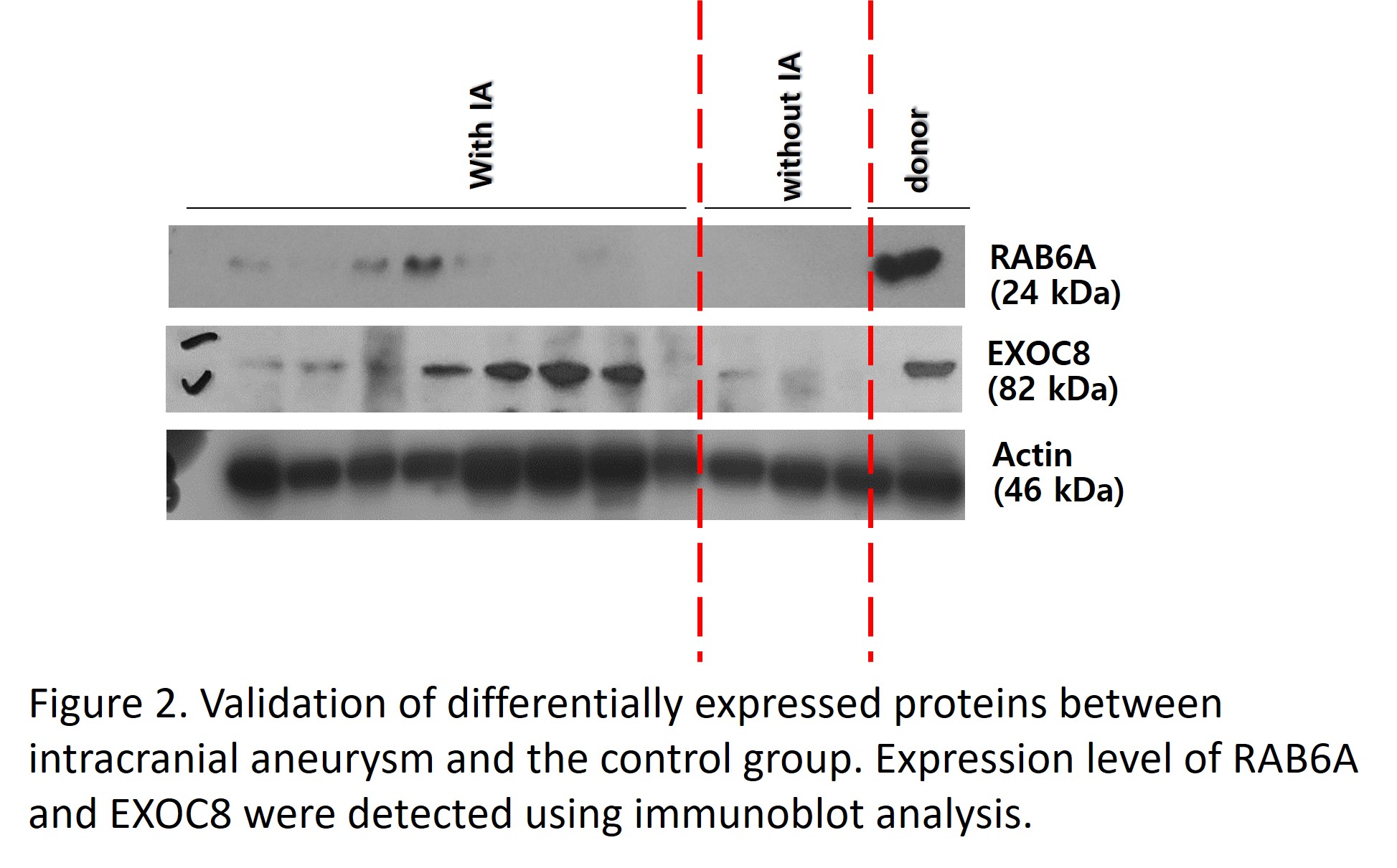
Results: Eight proteins showed significant variation between IA and control groups, with four proteins upregulated (DIS3, MMS19, EXOC8, RAB6A) and four downregulated (CLUH, SYNC, MEF2D, WDR36) in IA group (Log2 fold change (FC) >2 and false discovery rate [FDR] q-value <0.05) compared to the control group. These proteins correlated with pathways implicated in IA development, such as ciliopathy, exocytosis, inflammation, extracellular matrix remodelling, and apoptosis. These proteins were quantitatively validated using immunoblot and found to be consistent with proteomic data. Moreover, a connection was observed between protein expression and clinical metrics (bilirubin, prothrombin time, platelet count), indicating their potential as early diagnostic markers.
Conclusion: This study is the first to employ renal artery samples to study underlying mechanisms for IA in ADPKD patients by proteomics. We identified and validated novel candidate markers that are either upregulated or downregulated in the IA group compared to the control group. This research's finding opens new avenues for understanding and diagnosing IA in ADPKD, potentially leading to earlier diagnosis and targeted treatments.