Machine learning in transplant medicine: Predicting diarrheic diseases in kidney recipients
Alejandro Camargo5, Liceth Castañeda1, Carlos Puentes2,3, Alejandro Duitama2,3, Andrés Cardona2,4, Consuelo Romero2,4, Alejandro Ramos2,4, Sandra Perdomo2,4, Andrea Garcia-Lopez5, Andrea Gomez-Montero5, Fernando Giron-Luque5.
1Magíster en Estadística Aplicada y Ciencia de Datos, Universidad El Bosque , Bogota, Colombia; 2Laboratorio de Inteligencia Artificial , Universidad El Bosque , Bogota, Colombia; 3Grupo SIGNOS, Universidad El Bosque , Bogota, Colombia; 4Grupo de Inmunología Celular y Molecular , Universidad El Bosque , Bogota, Colombia; 5Colombiana de Trasplantes , Bogota, Colombia
Introduction: Diarrheic diseases represent a relevant post-transplant complication for kidney transplant recipients, often leading to adverse outcomes that can compromise graft function and patient well-being. The complexity of managing such conditions is heightened by the intricate balance required in immunosuppressive therapy, which can predispose patients to infectious and non-infectious causes of diarrhea. Early and accurate prediction of diarrheic diseases is critical for timely intervention, potentially improving patient management and outcomes. This study presents a comparison of machine learning models to predict acute diarrheal disease (ADD) in patients with kidney transplantation in Colombia.
Methods: A retrospective cohort of 622 kidney transplant recipients followed for one year at Colombiana de Trasplantes were analyzed. A total of 1353 records were classified into two groups ADD (Yes/No). Specific data imputation techniques were applied by class to complete missing data coherently within each group. Oversampling was used to address data imbalance, especially in the minority class. Variable selection was performed using SelectKbest technique. Comparisons between 15 models were performed using confusion matrix, sensitivity, specificity, and AUC. All models underwent cross-validation on normalized data.
Results: Models with higher performance were the Random Forest Classifier (AUC 96.1%), the Extreme Gradient Boosting (AUC 95.3%), and Light Gradient Boosting Machine (95.1%). It is highlighted that the Random Forest Classifier model showed the best metrics, confirmed by the confusion matrix, where sensitivity and specificity for test data are 77% and 82%. Influential variables in this prediction included transplant to ADD time, Mycophenolate Mofetil, Mycophenolate Sodium, Deflazacort, Hypertension, Diabetes, Retransplant, Donor Type, and gender.
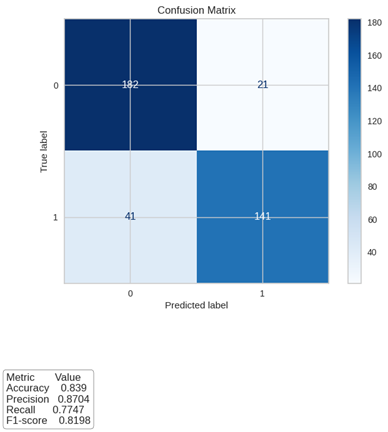
Conclusions: The final model demonstrated strong performance in predicting diarrheic diseases among kidney transplant recipients. While it represents a step forward in leveraging machine learning to address a critical post-transplant complication, the findings underscore the need for further refinement and validation of the model. This study highlights the potential of predictive analytics in improving post-transplant care, yet also emphasizes the challenges in developing highly accurate models for complex clinical conditions.
[1] Machine Learning
[2] Clinical Prediction Model
[3] Kidney Transplantation
[4] Diarrhea
