Interpretable prediction of kidney allograft rejection using machine learning: A comparison between linear and non-linear models
Alvaro Assis de Souza1, Andrew P. Stubbs2, Dennis A. Hesselink1, Carla C. Baan1, Karin Boer1.
1Internal Medicine, Erasmus Medical Center, Rotterdam, Netherlands; 2Pathology and Clinical Bioinformatics, Erasmus Medical Center, Rotterdam, Netherlands
Introduction: Machine learning (ML) has been widely explored to predict allograft rejection, with comparable performance to traditional statistical methods. However, for implementing ML in clinical practice, understanding how these ‘black-box’ models arrive at their predictions is needed. This study uses interpretability techniques to compare the training of linear and non-linear models to predict the risk of kidney allograft rejection at the time of the first biopsy after kidney transplantation.
Methods: Data on donor kidney transplants, their recipients, and the transplantation procedure between August 2018 and December 2019 were included (N=130). The incidence of rejection at the first biopsy was 24%. Logistic regression with LASSO regularization and random forest with pre-specified hyperparameters were trained on six variables (occurrence of delayed graft function at day three post-transplant, donor body-mass index, donor age, donor type, warm ischemia time, and the performance of a preemptive transplant) to predict the probability of a rejection episode. The optimism-correct estimated performance of each model was based on a bootstrapping procedure (500x) and assessed by the area under the receiver operating characteristic curve (ROC-AUC), brier score loss, calibration curves, and net-benefit analysis. Shapley additive explanation (SHAP) and partial dependence plot (PDP) interpretability techniques were used to understand the models’ predictions.
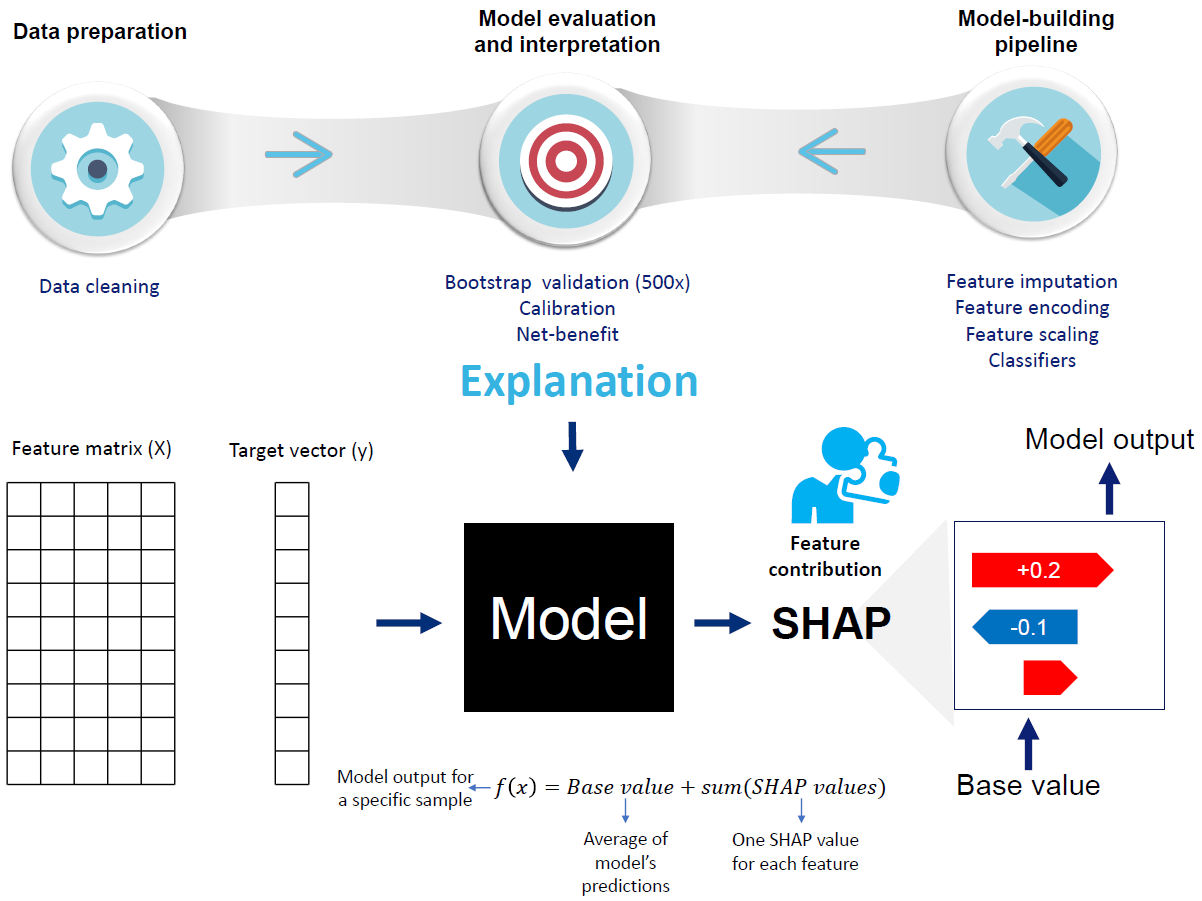
Results: Logistic regression and random forest models showed optimism-corrected ROC-AUC of 73% and 87%, respectively. The brier score losses, indicative of agreement between the predicted and observed probabilities, were 0.14 and 0.13 for the linear and non-linear models, respectively. The three most important features identified by SHAP (occurrence of delayed graft function at day three post-transplant, donor body-mass index, and warm ischemia time) agreed between logistic regression and random forest. PDP plots for the random forest revealed non-linear risk curves for rejection that are not naturally captured by logistic regression.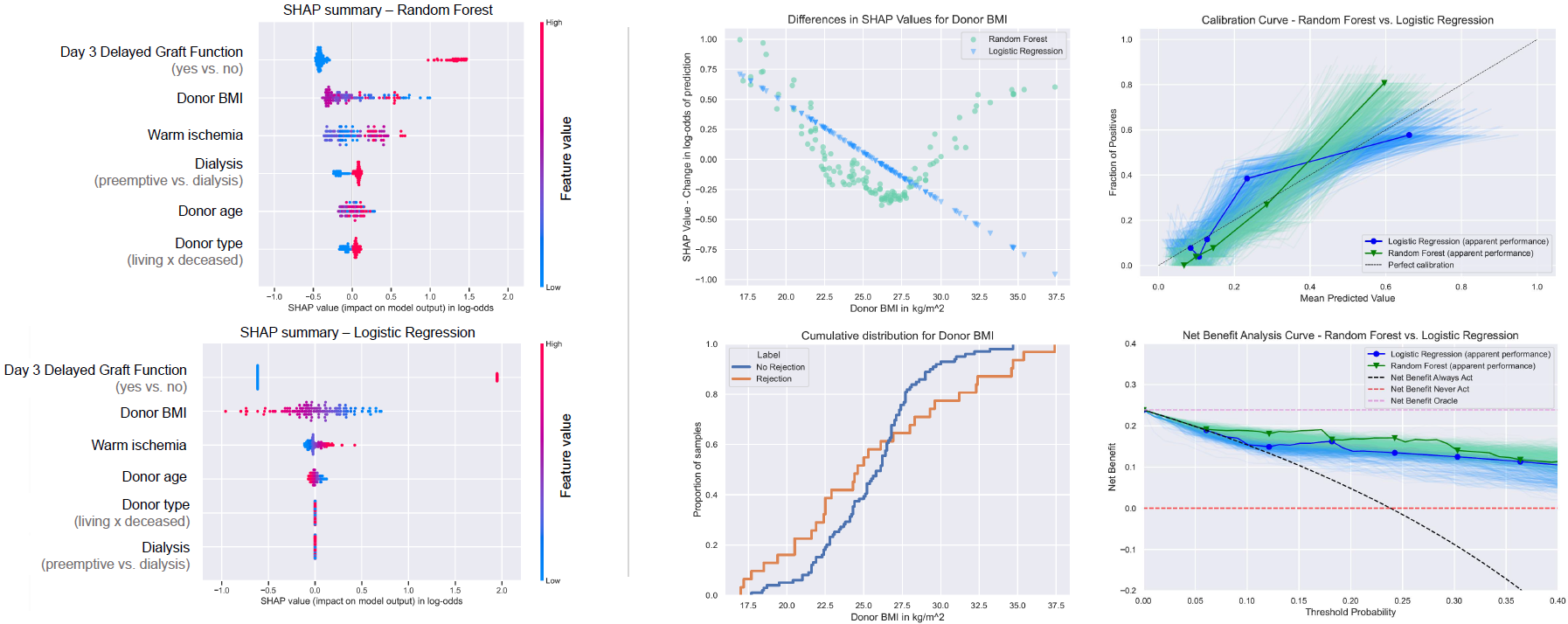
Conclusion: We extended the use of SHAP in predicting kidney allograft rejection. We showed differences in learning patterns between the linear and non-linear models that may have implications for medical decision-making. Our findings elucidate the need for interpretability in the development of predictive models for clinical practice.
Iacopo Cristoferi, MD.
[1] machine-learning
[2] kidney transplantation
[3] allograft rejection
[4] clinical prediction model
