Single-cell RNA sequencing to uncover differential expression genes using peripheral blood mononuclear cells according to rejection types in kidney transplant recipients
Jin-Myung Kim1, Seong Jun Lim1, Hye Eun Kwon1, Youngmin Ko1, Joo Hee Jung1, Hyunwook Kwon1, Young Hoon Kim1, Sung Shin1.
1Department of Surgery, Asan Medical Center, Seoul, Korea
Introduction: Despite the advancements in immunosuppressive medications, rejection remains the primary cause of early graft failure following kidney transplantation (KT). This underscores the necessity of gaining a deeper understanding of the composition and interactions within the alloreactive inflammatory infiltrate, which elucidates the distinctions between the two different types of rejections: antibody-mediated rejection (ABMR) and T-cell-mediated rejection (TCMR). In this study, we created profiles of various rejection types in peripheral blood mononuclear cells (PBMCs) using single-cell RNA sequencing and compared them with non-rejection biopsy samples to identify Differential Expression Genes (DEGs) associated with ABMR and TCMR.
Method: The study examined six kidney transplant recipients undergoing protocol biopsies, which were classified into three groups according to the Revised Banff 2019 criteria: No Major Abnormality (NOMOA, control), TCMR, and ABMR. The study used comprehensive single-cell RNA sequencing on PBMCs and applied Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to identify and annotate the functions of DEGs. The threshold was set as the adjusted p-value <0.05, and the absolute value of log-fold change | log2FC | ≥ 0.5 was statistically significant for the DEGs.
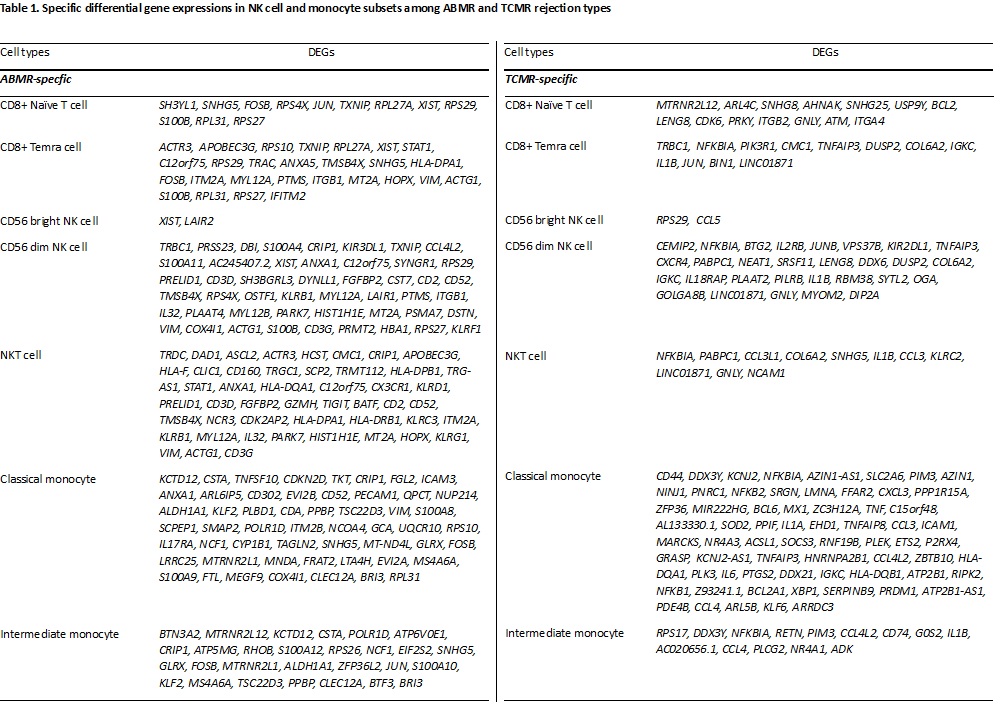
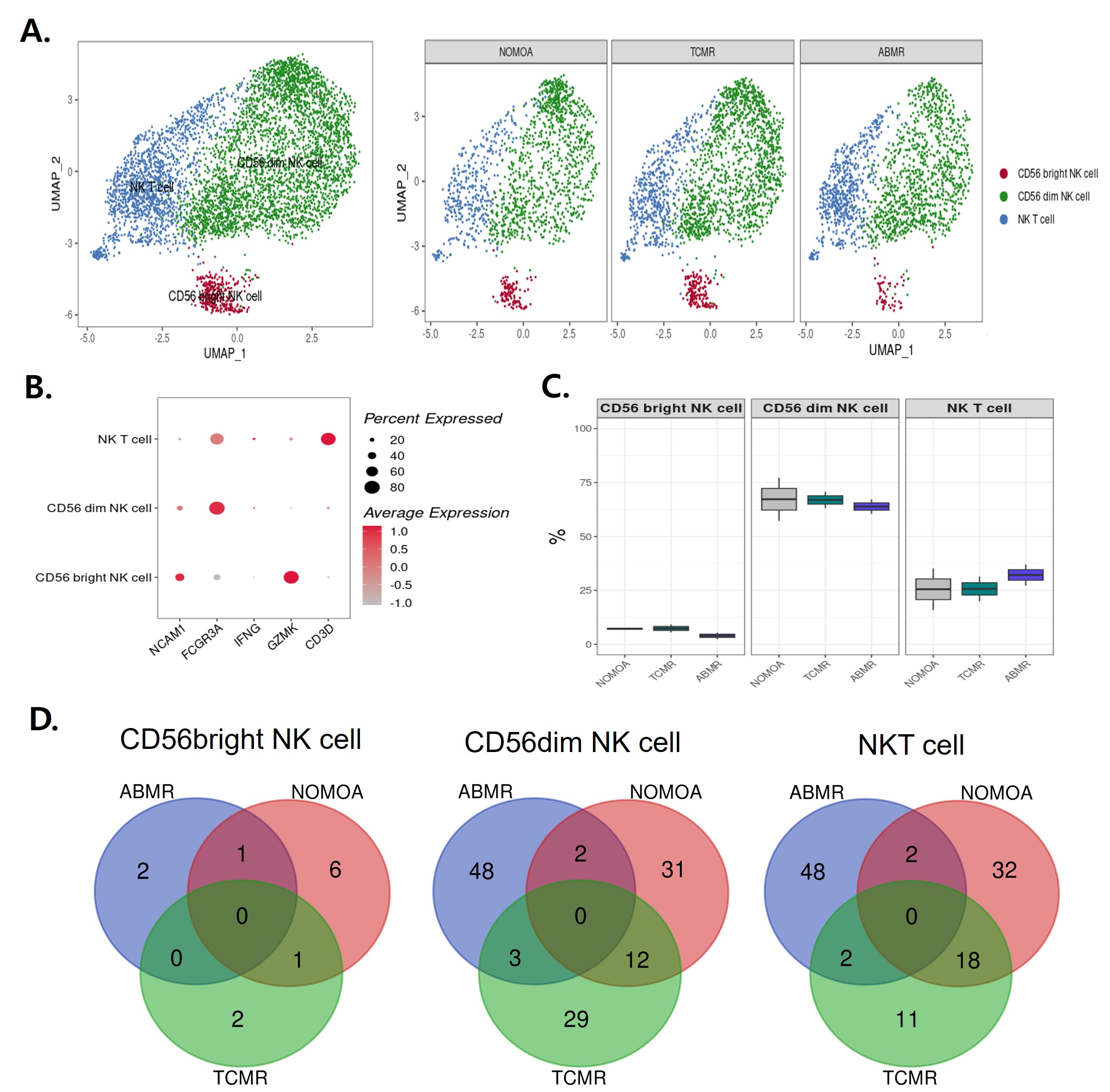
Results: The study unveiled distinctive cellular and genetic landscapes associated with ABMR and TCMR. Specifically, an enrichment of CD8+ Naive T cells and CD8+ terminally differentiated effector memory (TEMRA) T cells was observed in the ABMR group, with these cells expressing ABMR-specific DEGs including SH3YL1, JUN, and RPS4X. Monocytes were more distributed in ABMR than TCMR and showed 16 ABMR specific DEGs. In addition, while CD56bright NK cells in the TCMR group were linked to DEGs such as RPS29 and CCL5, CD56dim NK cells demonstrated a unique set of 29 DEGs exclusively associated with ABMR, indicating profound differences in the immune responses between ABMR and TCMR.
Conclusion: The results from this study provided new insights into the cellular and molecular dynamics of kidney allograft rejection. By pinpointing specific cell types and DEGs linked to rejection types using single-cell RNA sequencing, the study opened new avenues for the non-invasive diagnosis and monitoring of allograft rejections in KT patients. These findings not only enhance our understanding of the immunological mechanisms behind rejection but also pave the way for the development of targeted therapies, potentially improving patient outcomes. Future investigations will be pivotal in validating these biomarkers and assessing their clinical relevance.
[1] graft rejection
[2] single-cell RNA sequencing
[3] differential expression genes
