Decoding the hallmarks of allograft dysfunction with a comprehensive pan-organ transcriptomic atlas
Harry Robertson1,2,3, Jennifer Li1,4, Elvira Jiminez-Vera1, Jean Yang2,3, Philip O'Connell1,4, Ellis Patrick2,3, Natasha Rogers1,4.
1Centre for Transplant and Renal Research, Westmead Institute for Medical Research, Westmead, Australia; 2School of Mathematics and Statistics, University of Sydney, Sydney, Australia; 3Sydney Precision Data Science Centre, University of Sydney, Sydney, Australia; 4Department of Renal Medicine, Westmead Hospital, Westmead, Australia
Introduction: The pathogenesis of allograft (dys)function has been studied using ‘-omics’ based technology. Advances in next-generation sequencing technology have unlocked new insights into molecular markers of disease. However, despite the extensive research into dysfunction mechanisms, a universal biopsy and blood-based biomarker of allograft rejection remains elusive. Furthermore, it is unclear whether this biomarker could be consistent across organs, or whether each organ requires individualised markers to identify dysfunction. Identifying such an organ-agnostic biomarker would significantly enhance personalized transplant care and improve patient outcomes.
Methods: We curated 150 public gene expression datasets at both bulk and single-cell resolution that examined transcriptomic changes in allograft rejection across heart, liver, lung, and kidney transplants. We established Transferable Omics Prediction (TOP) as the first transfer learning framework to construct AI models that leverage information across different organs. TOP was employed to model allograft rejection, in an organ-agnostic manner, from peripheral blood and biopsy samples, using 23 and 56 datasets respectively. Each model was internally validated using a leave-one-dataset-out cross validation strategy. These two models, representing organ-agnostic non-invasive and invasive diagnostic methods, were then validated using a single centre prospectively recruited cohort.
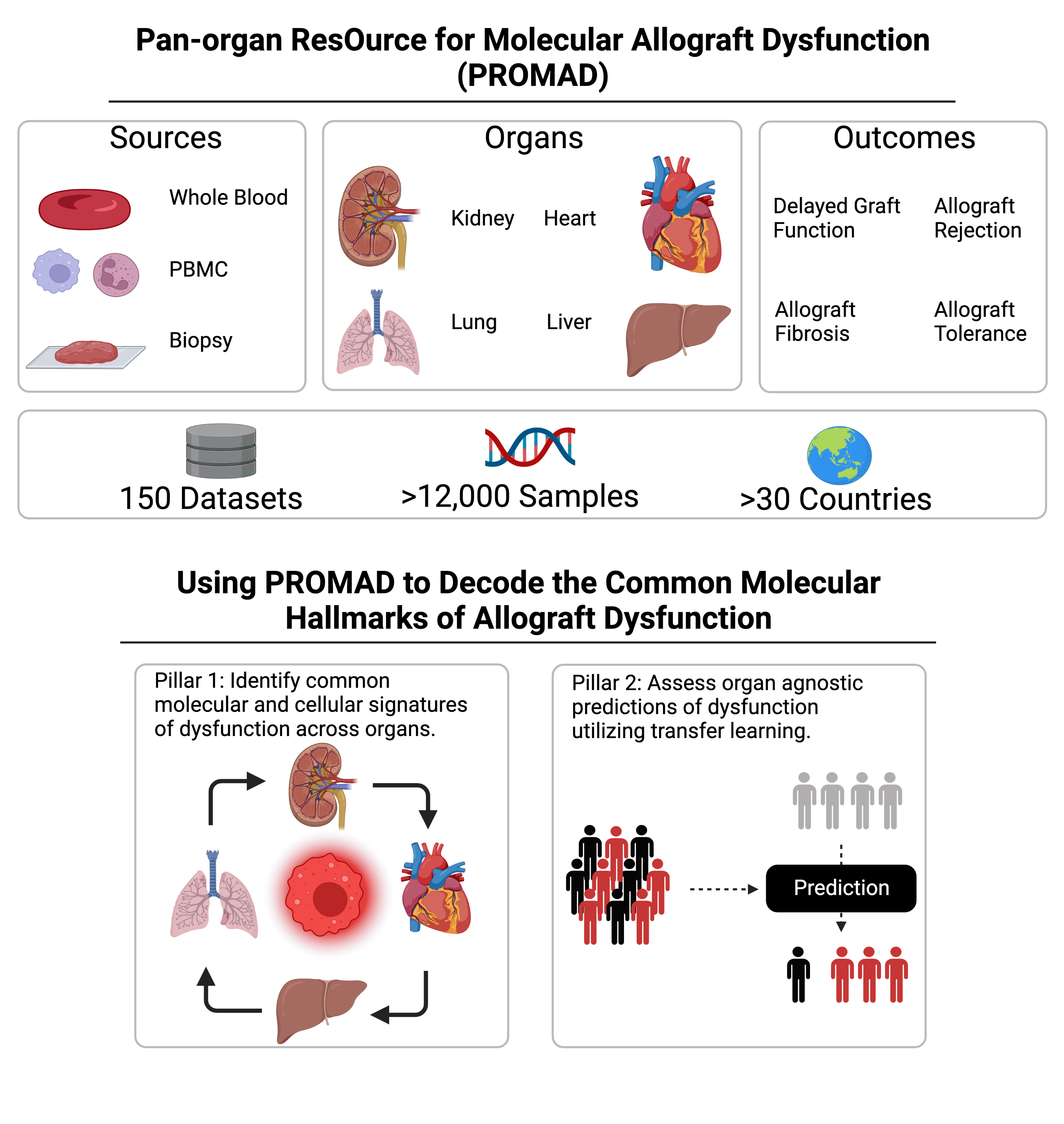
Results: We developed PROMAD, the first transcriptomic atlas in organ transplantation, through extensive curation of public datasets. Which has been made publicly available for the transplant community.
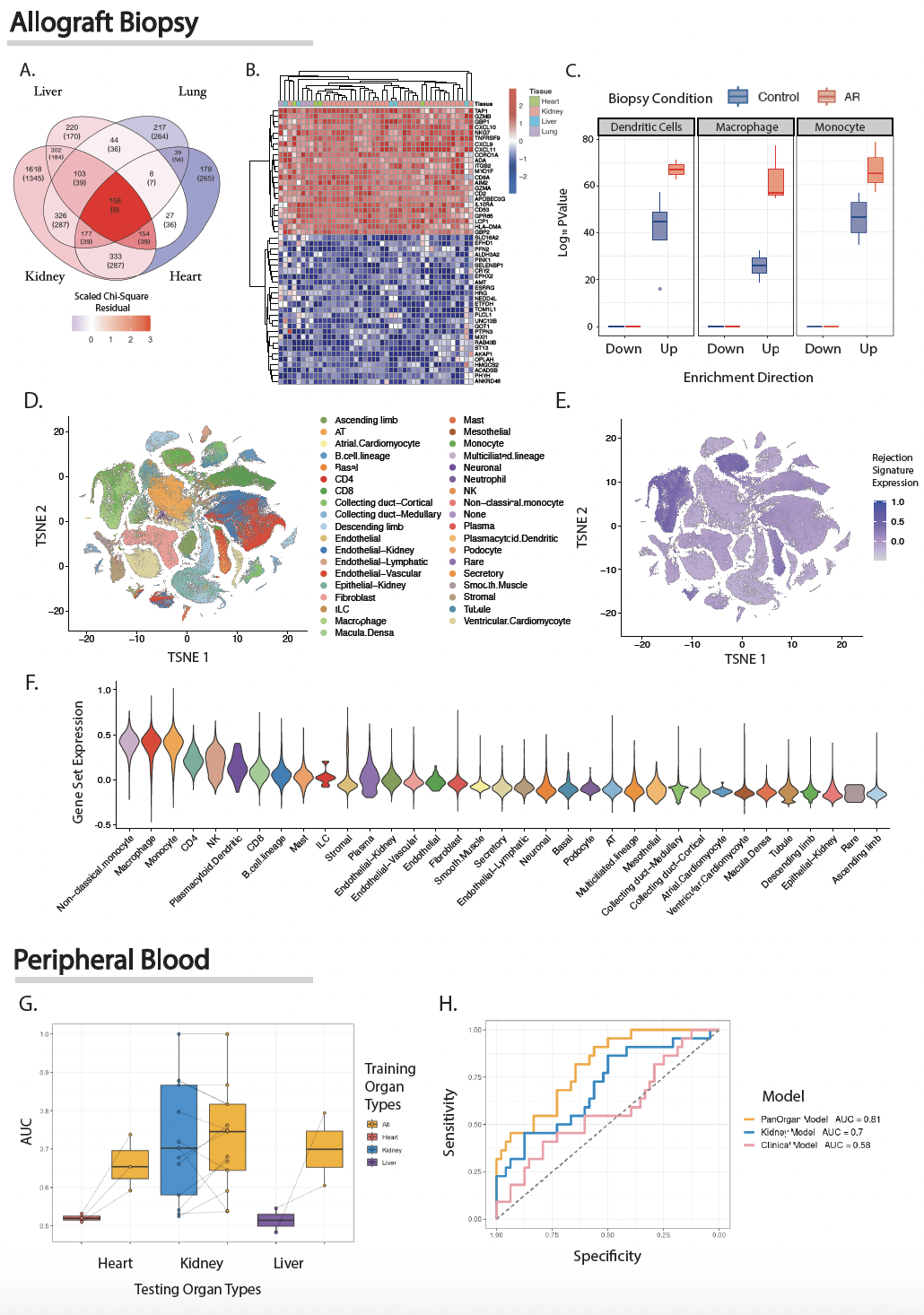
Integrating PROMAD using our novel machine learning method led us to develop two predictive models either biopsy or peripheral blood samples. Our biopsy model of allograft rejection demonstrated concordant transcriptomic features across different organs (Figure 2A-B). Implicating the same antigen presenting cell phenotype enriched across organs (Figure 2C-F).
Our pan-organ models, built from both invasive (biopsy) and non-invasive (peripheral blood) samples, showed significant diagnostic improvements over traditional organ specific biomarkers (Figure 2G). Validation in a prospective cohort revealed that our non-invasive biomarker outperformed the current clinical gold standard (Figure 2H).
Conclusion: We make extensive use of the PROMAD atlas to develop both an invasive and non-invasive method of detecting allograft rejection. Our study establishes the capacity for machine learning models to learn across organs, rationalizing the construction of a transcriptomic transplant atlas that can be employed to develop pan-organ biomarkers of allograft dysfunction. Through recruiting a cohort with comprehensive clinical profiling, we provide evidence supporting the clinical adoption of our novel, non-invasive diagnostic tool.
[1] Machine learning
[2] Omics
[3] Precision Medicine
[4] Graft Rejection
[5] Pan-Organ
[6] Transcriptomics
[7] Big data
