T cell receptors and the transcriptomic profile of t cells in transplant tolerance
Min Hu1,2, Juanjuan Zhang1, Hannah Wang1, David Bo Lu1, Elvira Jimenez-Vera1, Martina Raneri1, Paulomi Mehta1, Brian Gloss3, Harry Robertson1, Jennifer Li1, Wayne J Hawthorne1, Natasha Rogers1, Stephen I Alexander4, Philip J O’Connell1.
1Centre for Transplant and Renal Research, The Westmead Institute for Medical Research, Westmead, Australia; 2Faculty of Medicine and Health, The University of Sydney, Camperdown, NSW , Australia; 3Scientific Platforms, The Westmead Institute for Medical Research, Westmead, Australia; 4Centre for Kidney Research, The Children's Hospital at Westmead, Westmead, Australia
Introduction: We demonstrated that T-cell co-stimulatory blockade of the B7-CD28/CTLA4 and CD40-CD154 pathways using CTLA4-Fc and MR1 antibodies induced islet xenograft tolerance. We found that CD4+Foxp3+ regulatory T cells (Tregs) play an essential role for tolerance induction and maintenance in this model.
Aims: To investigate T cell receptors (TCRs) and the transcriptomic profile of T cells in transplant tolerance at a single cell level.
Methods: Foxp3GFP mouse-recipients were transplanted with pig islets and received a single dose of CTLA4-Fc (500 ug/mouse) at the day of transplantation and a dose of MR1 (500 ug/mouse) at days 0, 2, 4 and 6 to induce tolerance. After confirming islet-graft function by serum porcine C-peptide, at 100 days post-transplantation, samples of (1) spleen, (2) kidney-draining-lymph-node (DLN) and (3) grafts in tolerant-mice (n = 5); and (4) spleen and (5) DLN of control mice (without transplantation/treatment) (n = 5) were collected. One reaction/group of sorted CD3+ T cells containing 5 bio-samples with Totalseq anti-mouse hashtag antibodies was performed by10xChromium ScRNA-seq 5' gene expression (5 reactions in total).
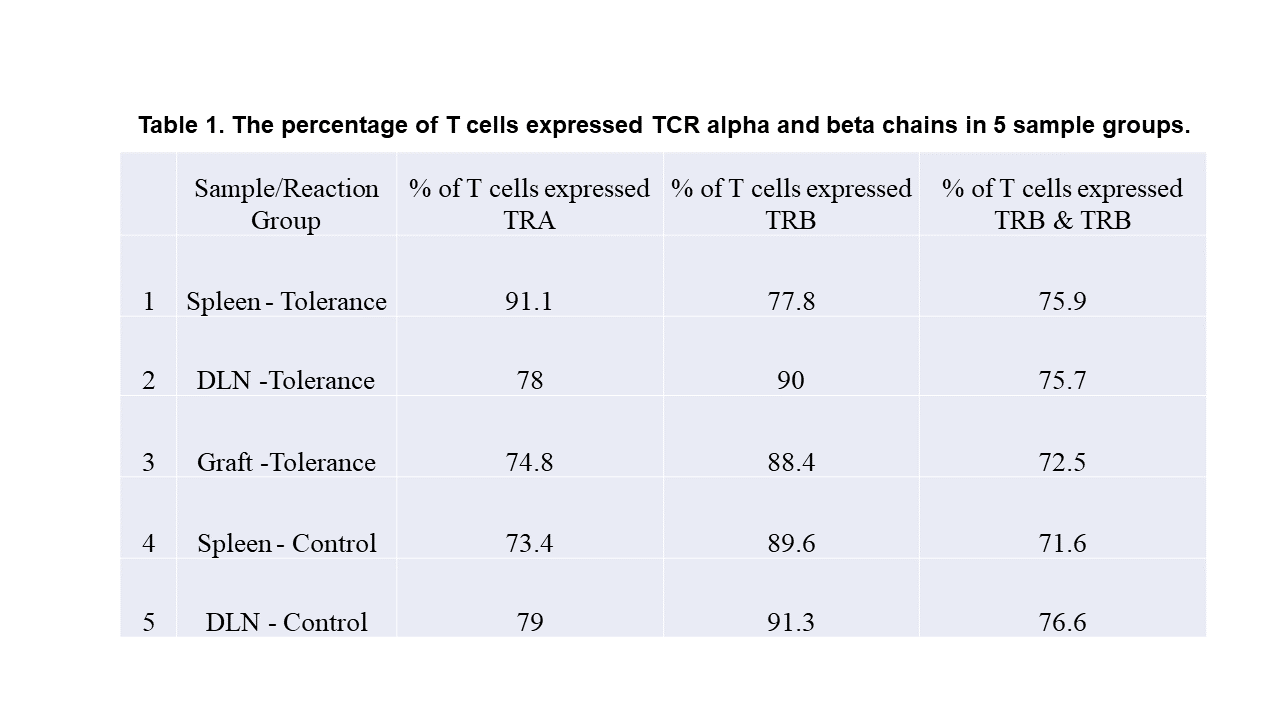
Results: 7621-16989 single cells/group passed QC filters and individual mouse cells within each group were identified. Transcriptomic profile showed 97%-99.7% of sorted cells were T cells containing naïve/resting and memory CD4+ and CD8+ T cells, NKT, CD8+Foxp3+, γδ T cells, and Tregs. A subset of cells expressed both TCR and B cell receptor was identified. A high Treg proportion was confirmed in tolerant mice. Interestingly, a high proportion of NKT cells (15 TRBV families) was observed in tolerant grafts. 74%-91.1% of T cells expressed TCR alpha (TRA), 77.8%-91.1% of T cells expressed TCR beta (TRB), and 71.6%-75.9% of T cells expressed both TRA and TRB (Table 1). While all 22 TRBV families were detected on Tregs of tolerant mice, a high proportion of Tregs that expressed TRBV29 was identified in both lymphoid organs and grafts of tolerant mice.
In conclusion: We established a method to evaluate TCRs and the transcriptomic profile of T cells in transplant tolerance at a single cell level and further analysis can be done for identification of key driven signalling and TCRs for antigen specific Tregs and memory T cells.
[1] Tolerance
[2] single cell RNA sequency
[3] T cell receptors
[4] transcriptomics
[5] CD4+Foxp3+ regulatory T cells
[6] T-cell co-stimulatory blockade
