Machine learning models for the early detection of urinary infections in post-kidney transplantation patients
Liceth Castañeda1, Carlos Puentes2,3, Alejandro Duitama2,3, Andrés Cardona2,4, Consuelo Romero2,4, Alejandro Ramos2,4, Sandra Perdomo2,4, Alejandro Camargo5, Andrea Gomez-Montero5, Andrea Garcia-Lopez5, Fernando Giron-Luque5.
1Magíster en Estadística Aplicada y Ciencia de Datos, Universidad El Bosque, Bogota, Colombia; 2Laboratorio de Inteligencia Artificial, Universidad El Bosque, Bogota, Colombia; 3Grupo SIGNOS, Universidad El Bosque, Bogota, Colombia; 4Grupo de Inmunología Celular y Molecular, Universidad El Bosque, Bogota, Colombia; 5Colombiana de Trasplantes, Bogota, Colombia
Introduction: Urinary tract infections (UTIs) are among the most common bacterial infections, posing significant health burdens globally. Early and accurate prediction of UTIs can significantly enhance patient outcomes, reduce healthcare costs, and minimize the risk of complications, including antibiotic resistance. This study presents a comparison of machine learning models to predict UTIs in patients with kidney transplantation within a Colombian population.
Methods: A retrospective cohort of 911 kidney transplant recipients followed for one year at Colombiana de Trasplantes were analyzed. A total of 29340 records were classified into two groups UTI Yes/No. Specific data imputation techniques were applied by class to complete missing data coherently within each group. Undersampling was used to address data imbalance, especially in the minority class. Variable selection was performed using featurewiz technique. Comparisons between 15 models were performed using confusion matrix, sensitivity, specificity, and AUC. All models underwent cross-validation on normalized data.
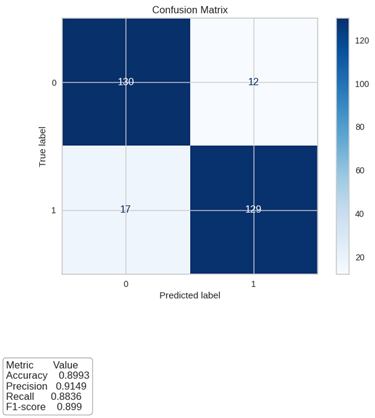
Results: Highest performance models were the Random Forest Classifier (AUC 95.6%) and the Extra Trees Classifier (AUC 94.2%). The Random Forest Classifier model showed the best metrics, confirmed by the confusion matrix, where sensitivity and specificity for test data are 88% and 89% respectively. Influential variables in this prediction included age, induction therapy, follow-up, body mass index, donor type, Mycophenolate Mofetil, diabetes, Tacrolimus, Mycophenolate Sodium, Prednisolone, Tacrolimus Levels, Everolimus, and Deflazacort.
Conclusion: The model demonstrated strong performance in predicting UTIs showcasing its potential as a valuable tool in clinical settings. With robust accuracy metrics, it provides a reliable basis for early identification and intervention, ultimately aiming to improve patient outcomes and reduce the burden of urinary tract infections. This promising result underscores the importance of further validating the model across diverse populations and clinical environments to ensure its efficacy and applicability in enhancing patient care.
[1] Kidney Transplantation
[2] Urinary tract infection
[3] Clinical prediction model
[4] Machine Learning
