Oliver McCallion, United Kingdom has been granted the TTS Scientific Congress Award
Spatial transcriptomics of kidney biopsies following adoptive regulatory T cell transfer reveals a marginal zone B cell signature and novel intra-graft immune interactions
Oliver McCallion1, Matthew Brook2, Paul Harden2, Joanna Hester1, Fadi Issa1.
1Nuffield Department of Surgical Sciences, University of Oxford, Oxford, United Kingdom; 2Department of Renal Medicine, Oxford University Hospitals NHS Foundation Trust, Oxford, United Kingdom
Introduction: Adoptive cell therapy (ACT) with autologous expanded regulatory T cells (Tregs) is a promising therapeutic strategy to enable immunosuppression minimisation in organ transplant recipients. The TWO study is the world’s largest phase II clinical trial of autologous polyclonal Treg ACT following living donor kidney transplantation currently being undertaken by our team. In Treg ACT treated patients there is interest in assessing the migration patterns of transferred Tregs and their impact on tissue alloresponses within the transplant.
Methods: We performed spatial transcriptomic profiling on six kidney transplant biopsies from four TWO study patients receiving standard-of-care immunosuppression (control, n=2) or Treg ACT (TR001, n=2) using FFPE samples on the 10X Genomics Xenium platform mapping 477 genes with immunofluorescence enabled cell segmentation. In addition, cryopreserved 38-week core biopsies from two control patients and one TR001 patient were enzymatically dissociated and CD45+ lymphocytes isolated by flow sorting. PBMC samples from 6 patients at 5 timepoints (2 control, 3 TR001) and Treg ACT single cell suspensions (n=3) were generated. Gene expression and V(D)J immune receptor libraries were constructed and sequenced for all samples.
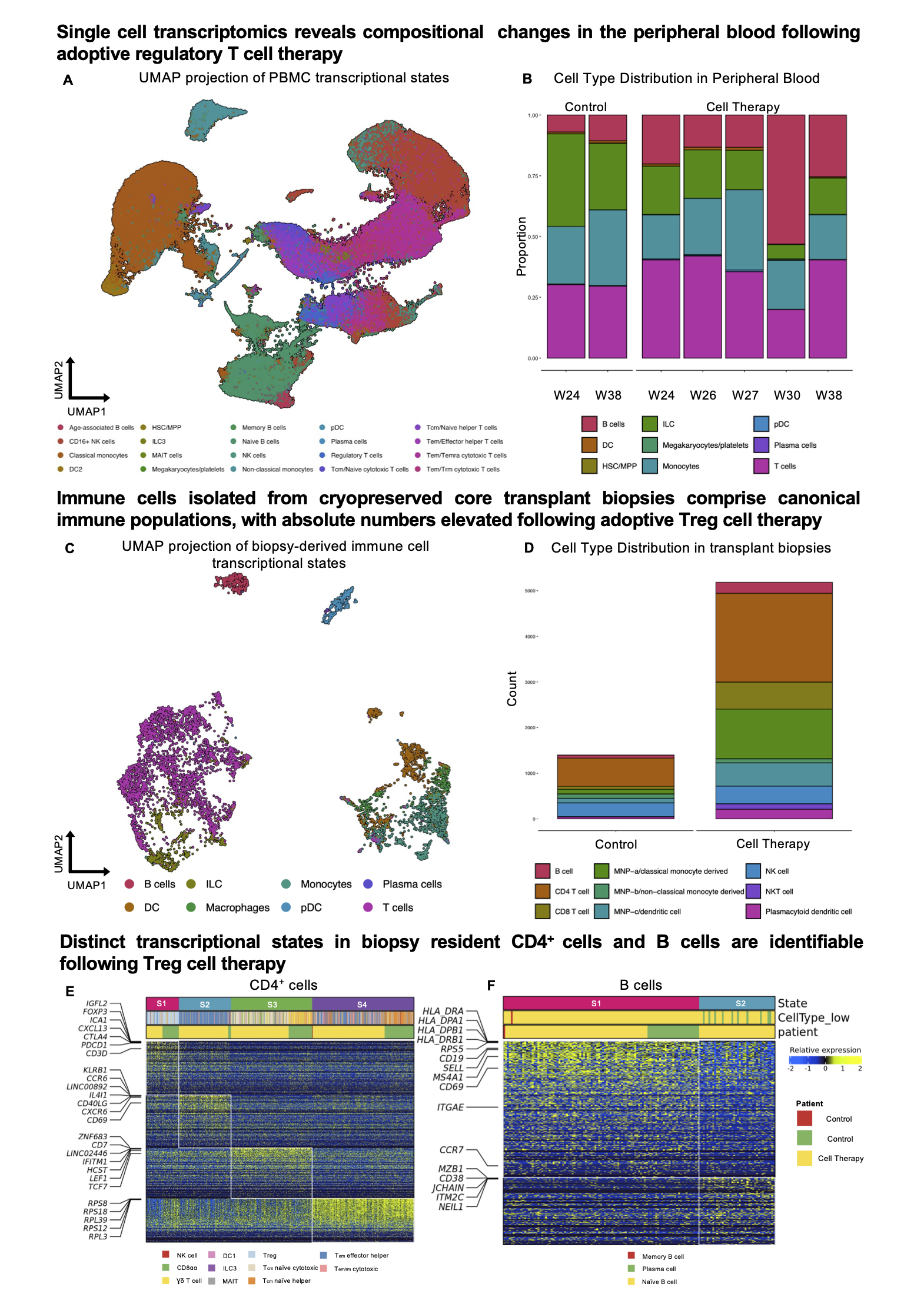
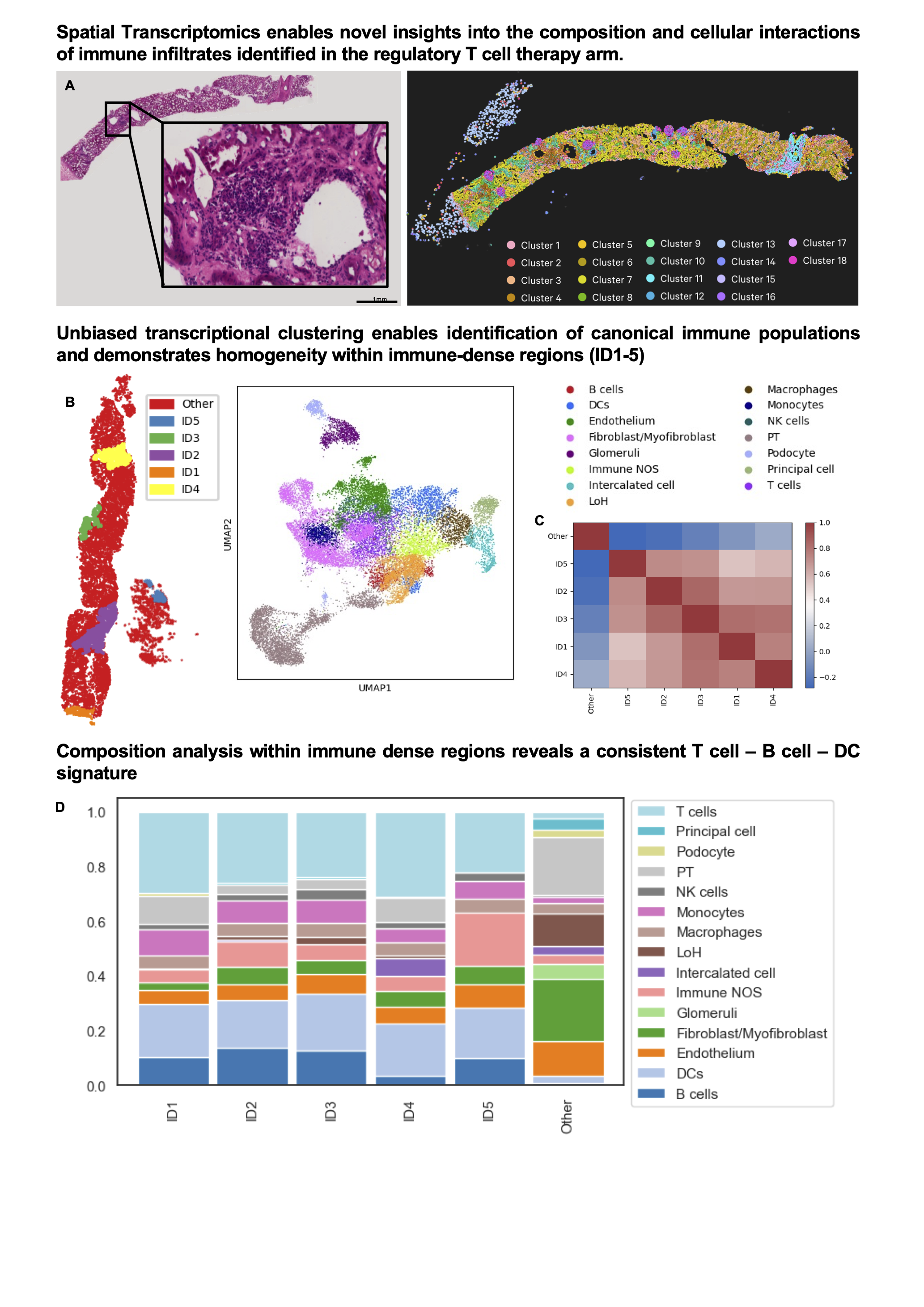
Results: A total of 6,500, 180,000, and 30,000 single cells from kidney biopsies, PBMC, and Treg ACT, respectively, passed QC filtering. Within peripheral blood, a proportional increase in B cells was observed following Treg adoptive transfer (Fig1. A, B). Immune cell recovery from the cell-therapy treated biopsy was significantly elevated and canonical lymphocyte populations were identifiable (Fig. 1C, D). Unsupervised evaluation of conserved and divergent transcriptional states between biopsies confirmed the presence of Tregs in both biopsies, characterised by a gene signature comprising FOXP3, CTLA4, PDCD1 and IGFL2 (Fig 1E). Interestingly a unique B cell state identified by the expression of MZB1, CD38, and JCHAIN indicative of marginal zone B cells was only identifiable within the biopsy of the Treg treated patient (Fig 1F). Spatial transcriptional profiling enabled the identification of renal structures and immune dense regions within transplant biopsies (Fig 2A, B). Immune dense regions (ID1-5) exhibited striking homogeneity (Fig 2C) and were comprised predominantly of T, B cell, and DC subpopulations (Fig 2D).
Conclusion: We identify a conserved Treg transcriptional signature within transplant biopsies from both standard-of-care and Treg-treated patients, additionally uncovering a marginal zone B cell subpopulation specific to the Treg treated biopsy. With cutting edge spatial transcriptomic approaches, we begin to define the composition and immune interactions characterising the alloresponse in patients receiving both conventional immunosuppressive therapy and adoptive Treg transfer.
Medical Research Council, Clinical Research Training Fellowship (MR/V000942/1) George Drexler Foundation Research Fellow, Royal College of Surgeons of England.
[1] Spatial Transcriptomics
[2] Single cell multiomics
[3] Tolerance
[4] Cell therapies
[5] Regulatory T cells
[6] Marginal Zone B cells
[7] Tissue alloresponses
