Sumoyee Basu, United Kingdom has been granted the TTS-FOCIS Basic and Translational Mentee-Mentor Award
OuTSMARTing donor specific antibodies (DSA): De novo DSA are preceded by a lack of specific regulation of CD4+ Th-1 Cells to DSA antigens but not other donor mismatches
Sumoyee Basu1, Dominic Stringer1, El Li Tham1, Chloe Martin2, Giovanna Lombardi1, Olivia Shaw2, Anthony Dorling1, Kate Bramham1.
1Centre for Urology, Nephrology and Transplantation, Kings College London, London, United Kingdom; 2Clinical Transplantation Laboratory, Guy's and St Thomas' NHS Foundation Trust, London, United Kingdom
OuTSMART Trial Investigators.
Introduction: While de novo donor specific antibodies (DSA) were associated with graft loss in the OuTSMART trial, optimised immunosuppression post-DSA failed to improve transplant survival. Furthermore it is poorly understood why some but not other patients even develop de novo DSA. This study therefore offered a unique opportunity to study the biology prior to DSA development given serial blood sampling of DSA- patients who then went on to develop de novo DSA during the study period. We evaluated concomitant changes in regulatory cells and IFNy and IL17 production, which knowingly associate with adverse transplant outcomes, germinal centre alloresponses and may pre-empt DSA formation.
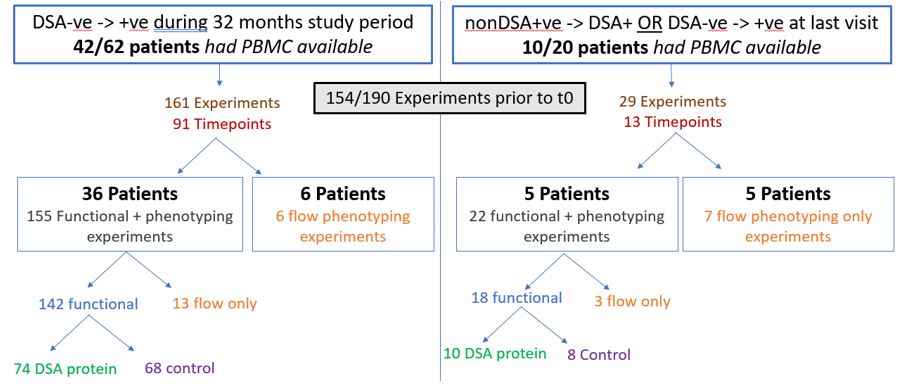
Methods: Peripheral blood mononuclear cells were available from multiple time points (Figure 1) prior to time of DSA development (t0) from 52/82 patients who were initially DSA- but developed DSA during the trial (Figure 1).
HLA Pure Proteins (PP) representing DSA or control donor mismatches were used to stimulate CD8 depleted PBMC to evaluate indirect allo-responses. Using FluoroSpot, antigen specific IFNγ and IL17 CD4+ production (ASR) was then compared, including conditions with additional depletion of CD19+ and CD25high cells.
Results: Antigen specific T cell responses (ASR) were detectable to either control or DSA PP in 67.9% (95/140) of the total experiments for IFNy and 35.7% (50/140) for IL17. Notably ASR were even measurable up to 32 months at all time points prior to t0 in more than 50% of samples.
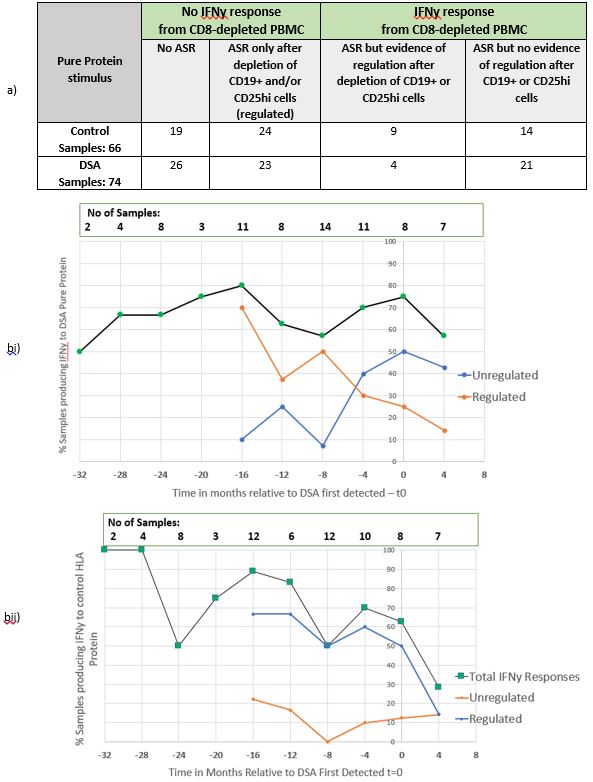
Figure 2a) demonstrates that overall ASR to control mismatched tended to be more regulated (50%, 33/66) than to DSA protein (36.5%, 27/74) but this was not statistically significant. However, Figure 2b) indicates that the proportion of regulated IFNy responses to both control and DSA PP were similar until 8 months prior to t0. However from -4 months, there was then a predominance of unregulated IFNy responses to DSA, which was not evident with the control protein where regulated responses were maintained. No similar patterns were discernible in responses to IL-17.
Conclusion: This is the first systematic study of clinical samples prior to de novo DSA development. This shows detectable T cell sensitisation even 32 months prior to DSA formation in more than 50% samples. Furthermore there was differential loss of regulation of IFNy production to DSA antigens just prior to DSA first detection. Comparatively control ASR had a predominance of more regulated responses at all time points. Ongoing deep phenotyping flow cytometry analyses will reveal the regulatory cells responsible and predict those who develop DSA. This work highlights a potential and a crucial window of detectable T cell sensitisation prior to de novo DSA which may be targeted with specific Treg cell therapy.
Medical Research Council. Guy's & St.Thomas' Kidney Patients' Association. National Institute for Health Research Efficacy and Mechanism Evaluation programme grant (ref 11/100/34).
[1] De novo DSA
[2] Allograft survival
[3] Sensitization
[4] Allorecognition
