Utilization of AlloView integrative dd-cfDNA model in predicting the risk of kidney allograft rejection
Sapna V Shah1, Santhi Voora1, Paul Hanson2, Wenlan Tian2, Shree Patel2, Thin Thin Maw1.
1Division of Nephrology and Hypertension, University of Southern California, Keck School of Medicine, Los Angeles, CA, United States; 2CareDx, Brisbane, CA, United States
Introduction: AlloView is the first AI-derived integrative dd-cfDNA (idd-cfDNA) model to predict rejection risk in kidney transplant recipients (KTR) by integrating dd-cfDNA (AlloSure®) and other clinical factors independently associated with rejection including eGFR, graft instability, prior rejection, and anti-HLA DSA. The discrimination performance of AlloView has been validated in a large, multi-center, international cohort. Compared to standard of care (SOC), the integrative model could provide clinicians with a more standardized, objective, and multidimensional assessment tool to discriminate rejection in KTR. However, a paucity of data describes a reference range to guide clinicians while interpreting AlloView results and determining whether to perform a biopsy. For the first time, we compared AlloView results by histological diagnosis.
Method: KTR from 1/1/2017 to 12/31/2023 with at least one biopsy, dd-cfDNA result ≤30 days preceding biopsy, and all AlloView clinical SOC input variables were included. Patients with BK virus nephropathy (BKVAN) or pyelonephritis were excluded. Primary analysis compared median AlloView results stratified by histological diagnosis: acute cellular rejection (ACR), acute antibody-mediated rejection or mixed rejection (AMR), borderline rejection, and no rejection. No rejection cohort also included acute tubular injury/necrosis, glomerulopathy (i.e., glomerular disease, such as FSGS, membranous nephropathy) and calcineurin inhibitor toxicity. Nonparametric tests were used to assess categorical and numerical variables. The area under the receiver-operating characteristic curve (AUROC) was used to determine the discriminating accuracy of AlloView.
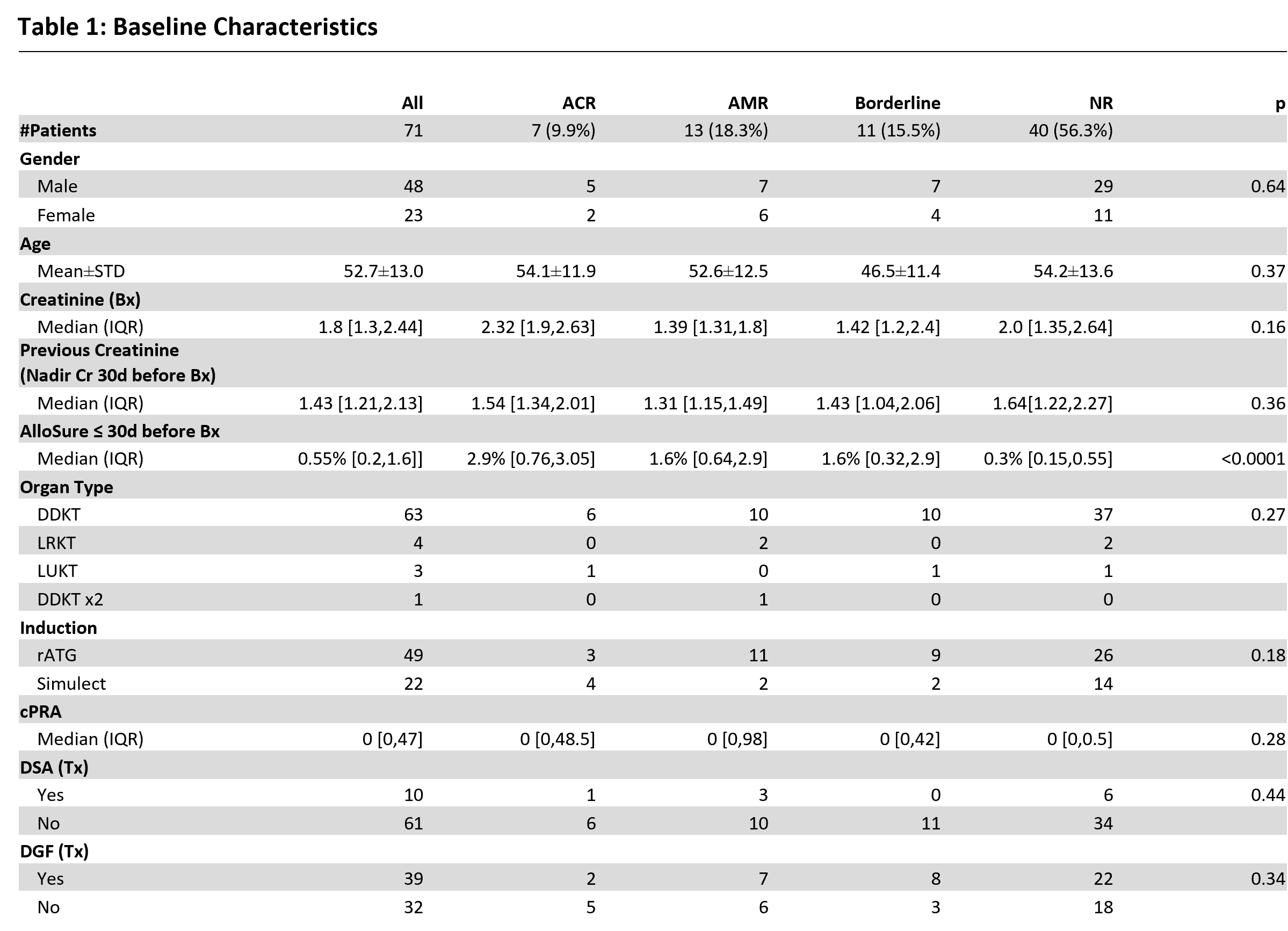
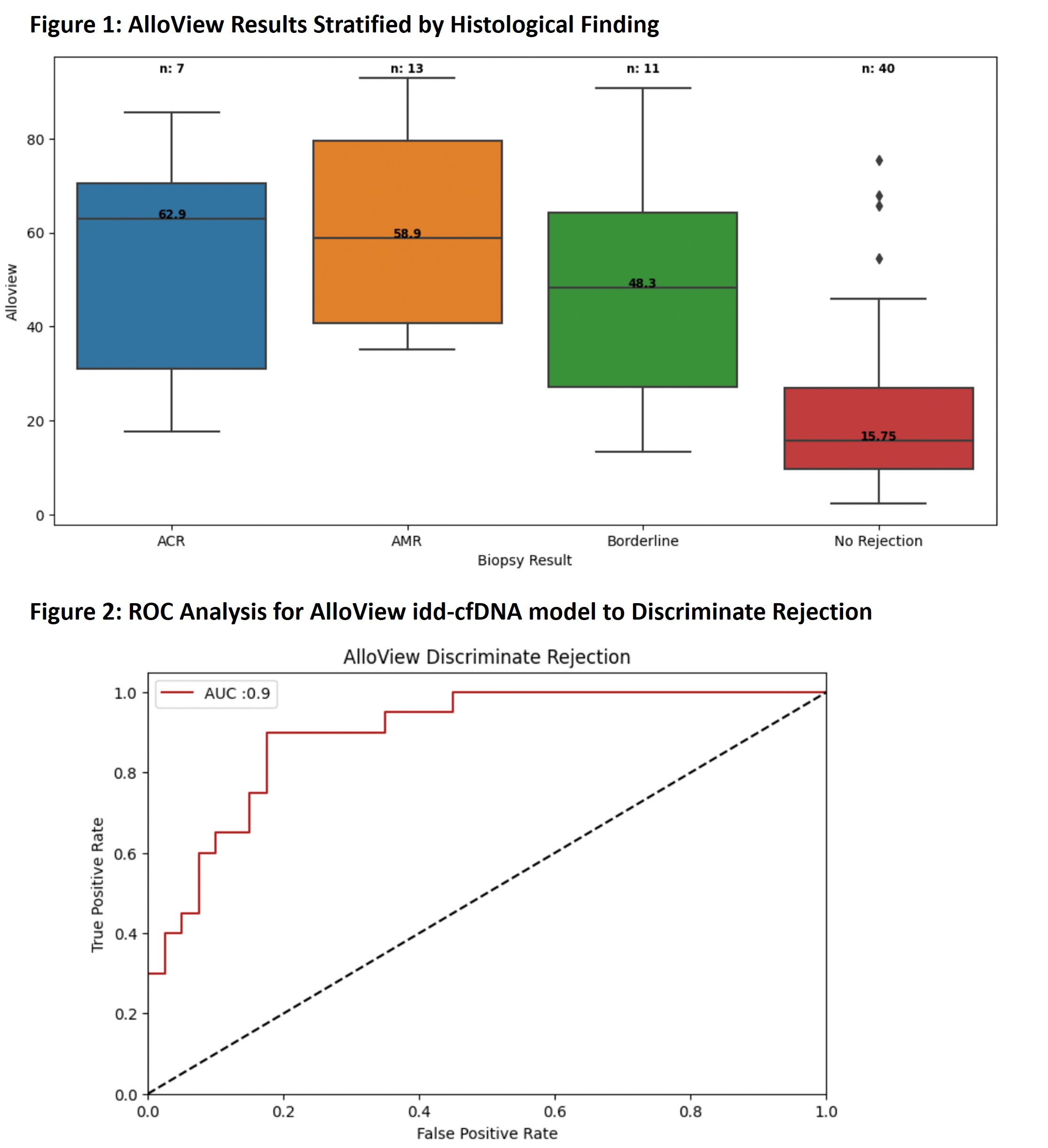
Results: A total of 71 AlloView results from 71 KTR met criteria for analysis. Most baseline characteristics had no significant difference between groups, with the exception of the biopsy-paired AlloSure results [Table 1]. There were 7 biopsies with ACR, 13 AMR events, 11 borderline rejections, and 40 no rejections. Median AlloView score was significantly higher in both ACR (62.9%, IQR 31.0-70.6%) and AMR (58.9%, IQR 40.8-79.5%) compared to no rejection (15.75%, IQR 9.65-26.9%) (p=0.0 and p=0.0037, respectively) [Figure 1]. Borderline rejection yielded a median AlloView of 48.3% (IQR 27.2-64.3%), which was also significantly different than no rejection (p=0.0016). The AUCROC for AlloView to discriminate rejection was 0.9 (95% CI: 0.82-0.98) [Figure 2].
Conclusion: Our data highlights for the first time the median AlloView values in rejection versus no rejection. The significant differences seen highlight the utility of AlloView in discriminating patient individual risk of rejection. With further validation, these references may support interpretation of the model.
[1] Kidney, transplant, Artificial Intelligence, machine learning, diagnostics, predictive modeling
