Single-cell RNA sequencing provides insights into intragraft B cell complexity in chronic lung allograft dysfunction (CLAD)
Ankita Burman1, Allen Duong1, Sajad Moshkelgosha1, Ke F Bei1, Stephen Juvet1, Tereza Martinu1.
1Toronto Lung Transplant Program, Latner Thoracic Research Laboratories, University Health Network, University of Toronto, Toronto, ON, Canada
Rationale: Chronic lung allograft dysfunction (CLAD) is the major factor limiting long-term survival in recipients of lung transplantation. CLAD has two clinical phenotypes – bronchiolitis obliterans syndrome (BOS) and the more severe restrictive allograft syndrome (RAS). We hypothesized that B cells in the CLAD lung allograft are a heterogeneous population with a unique composition and transcriptional signature, distinct from circulating B cells and from healthy lung B cells.
Objective: We aimed to comprehensively define the transcriptomic landscape of B cells in CLAD lung allografts explanted at re-transplantation, and to further delineate B cell differences between BOS and RAS phenotypes.
Methods: Single cell suspensions were made from CLAD lungs and matched peripheral blood mononuclear cells (PBMCs) (n = 13 comprising of 6 BOS and 7 RAS), and normal lungs (n = 6). All cells were subjected to single-cell RNA sequencing (scRNAseq) using the 10X Genomics 3’ assay. Furthermore, B and T cells were sorted from normal lungs (n = 4), CLAD lungs and matched PBMCs (n = 6 comprising of 3 BOS and 3 RAS); these underwent scRNAseq using the 10X Genomics 5’ assay. The 3’ data were utilized to calculate relative B cell proportions. The 5’ data were utilized to analyze B cell transcriptional heterogeneity using Seurat.
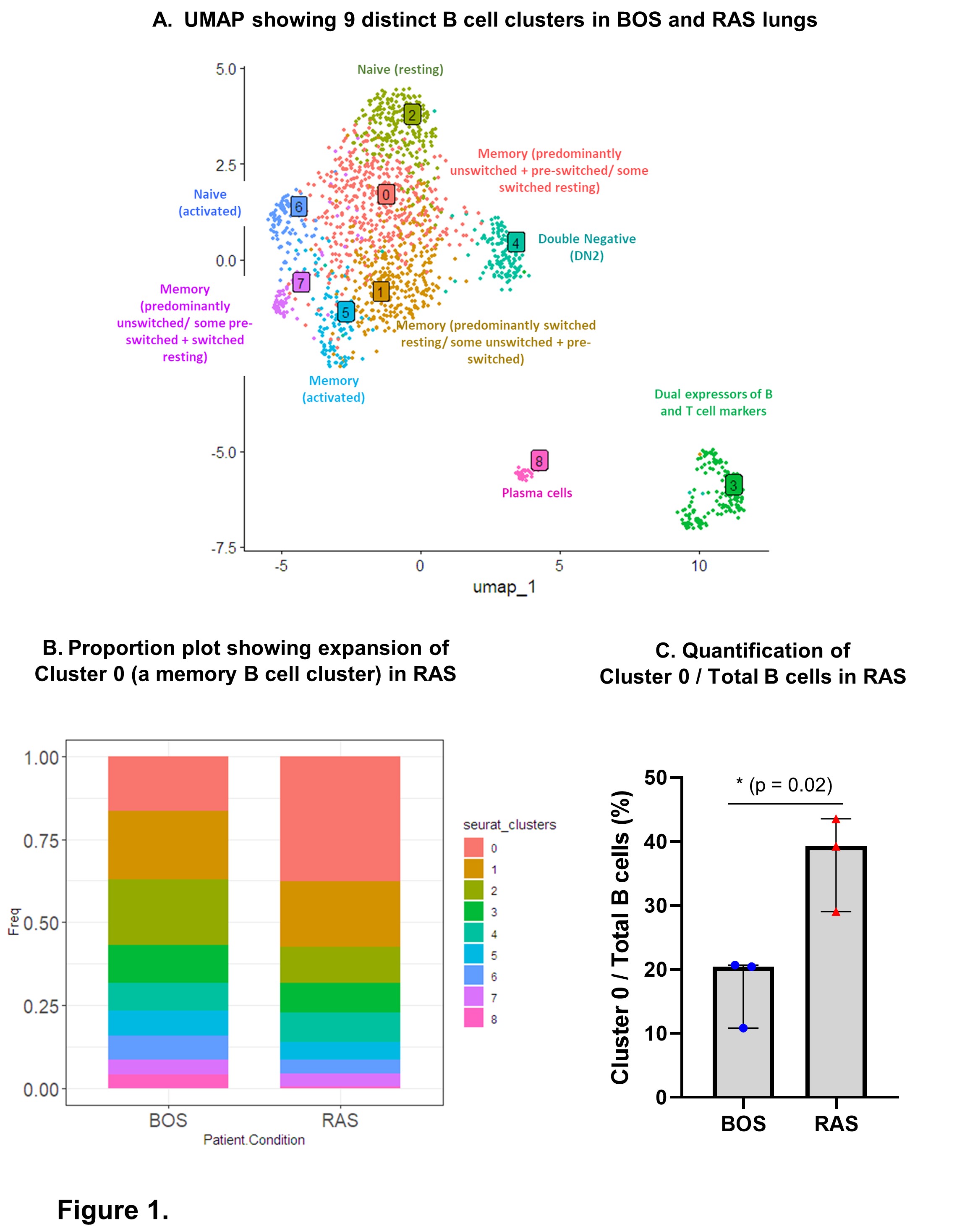
Results: Immune cells (CD45+/Total) and B cells (CD20+ CD19+ CD79A+/Total) were increased in CLAD compared to normal lungs. Transcriptomic analysis revealed 11 distinct B cell clusters in CLAD and normal lungs, with pathways related to lymphocyte activation and antigen processing/presentation upregulated in CLAD lung B cells compared to normal lung B cells. We found that the relative proportion of a naive B cell cluster was increased, and a CD27- IgD- B cell cluster was decreased, in CLAD lungs compared to matched PBMCs. We found a non-statistically significant increase in the proportion of B cells (B cells/CD45+ cells in Lungs/PBMC) in RAS compared to BOS patients. Expansion of a specific memory B cell subpopulation was observed in RAS compared to BOS lungs (Figure 1).
Conclusions: Our study deconvolutes B cell heterogeneity, the relative proportions of various B cell subpopulations, and the unique transcriptomic profiles of B cells in lung allografts of BOS and RAS patients. Our findings may help future identification of alloreactive B cell subsets, which could potentially be targeted for prevention or treatment of CLAD.
CIHR Project Grant 173343 (SJ, TM), CME Fellowship in Respiratory Medicine (AB), Sanofi innovations award (TM, SJ).
