Multi-omic profiling in a human decedent receiving a pig kidney xenograft reveals concerted immune responses preceding a biopsy confirmed rejection event
Brendan Keating1, Karen Khalil1, Jacqueline Kim1, Ian Jaffe1, Marc Lorber3, David Ayares2, Vasishta Tatapudi1, Jeffrey Stern1, Adam Griesemer1, Robert A Montgomery1.
1Surgery, NYU Langone Transplant Institute, New York, NY, United States; 2Revivicor Inc, , Blacksburg, VA, United States; 3United Therapeutics, Silver Spring, MD, United States
Introduction: Transplanting pig xenografts into human decedents affords a unique opportunity to deeply profile and analyze longitudinal blood and tissue timepoints. Recent advances in single-cell RNA sequencing (scRNAseq) and proteomic technologies afford unparalleled abilities to deeply profile blood and tissue in an unbiased (untargeted) fashion.
Methods: Daily blood samples were obtained from a human decedent receiving a pig kidney xenograft procedure lasting 61 days, with biopsies available over 9 timepoints including a biopsy confirmed antibody mediated rejection (AbMR) event on post-operative day (POD) 33. We performed proteomic screening using the Proteograph XT (Seer, CA) on 63 timepoints along with scRNAseq, B- and T-cell repertoire seq, and bulk RNAseq on 30 PBMC timepoints, and in situ geospatial transcriptomics on 9 biopsy timepoints (Xenium, 10x genomics), to assess and integrate the molecular signatures across the rejection event.
Results: Over 6,870, and 1,850 proteins were assignable to the human and pig proteomes respectively, for each of the blood plasma samples. Over 90% of all complement pathway proteins were detectable and their longitudinal signatures correlate with the AbMR and the subsequent successful treatment including pheresis and complement inhibitor treatments. scRNAseq show a concerted immune-cell response preceding AbMR and the subsequent treatment, as illustrated in Figure 1. Xenium data showed >20% of all cells in the POD 33 biopsy timepoint are human infiltrating immune cells, with no human cells evident in earlier timepoints, with a sharp drop-off of human cells after POD 33 AbMR treatment.
Conclusion: Multiomic profiling of blood and tissue samples over 61 days of a human decedent receiving a pig kidney xenograft reveals yields a number of immunological insights into a successfully treated AbMR episode in the longest pig-to-human xenotransplantation ever conducted.
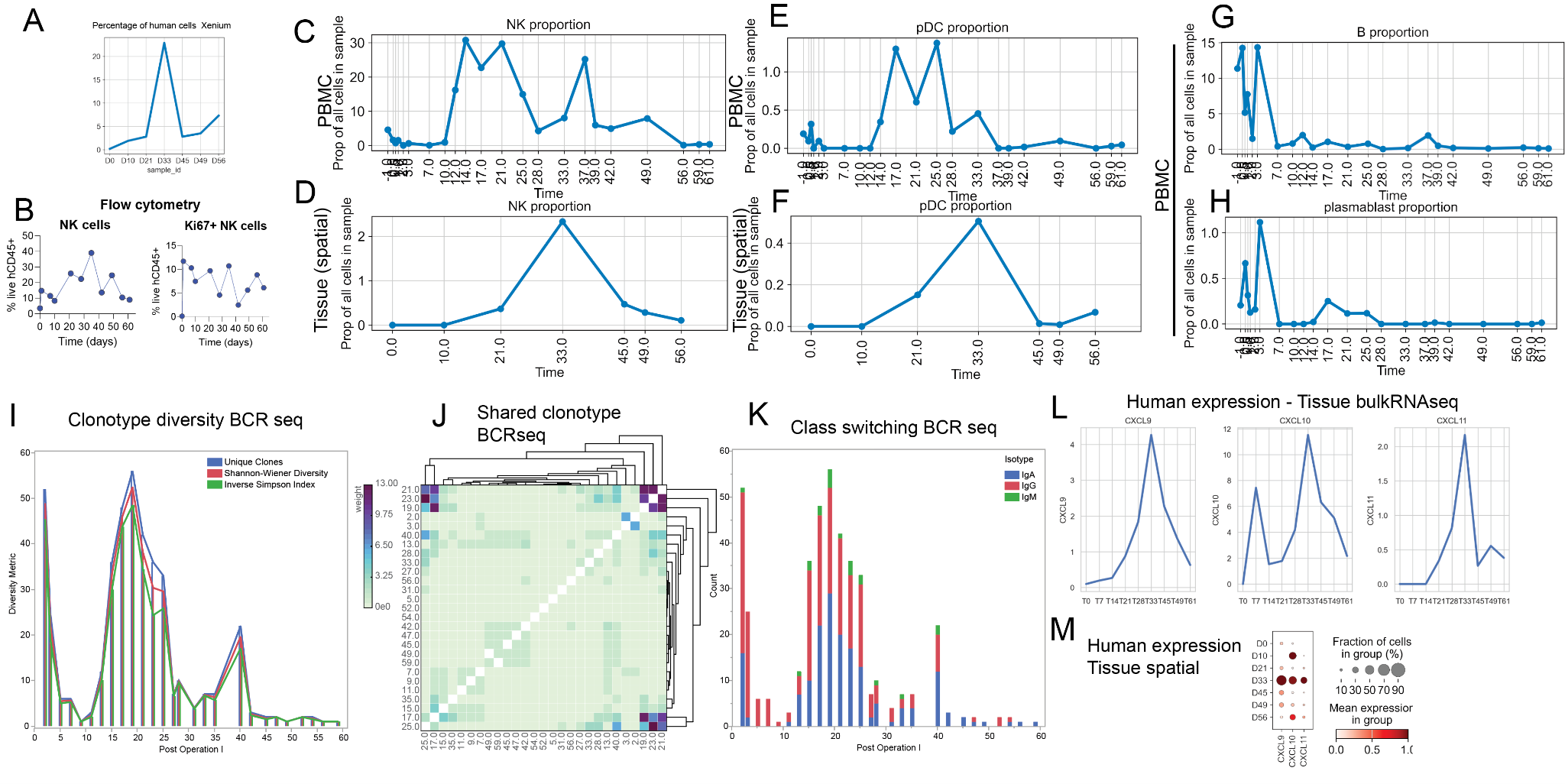
Figure 1: Early host immune cell response involving NK cells, pDCs and plasma / B cells. A. Percentage of human cells identified on the spatial transcriptomics Xenium slide, across all identified cells. B-D. Percentage of NK cells observed on B: Flow cytometry, C: PBMC scRNA-seq (out of all cells) and D: Spatial transcriptomics. E-F. Percentage of NK cells observed on E: PBMC scRNA-seq (out of all cells) and F: Spatial transcriptomics. G-H: Percentage of B cells (G) and plasmablasts (H) observed in PBMC scRNA-seq. I. Clonotype diversity from PBMC BCRseq J. Shared clonotypes across timepoints for PBMC BCRseq. L. Expression of human CXCL9, CXCL10, CXCL11 observed in the tissue bulk RNAseq. M. Expression of human CXCL9, CXCL10, CXCL11 observed in the tissue spatial transcriptomics.
