Multi-omics profiling in human decedents receiving genetically modified pig hearts reveals hallmarks of perioperative cardiac xenograft dysfunction
Brendan Keating1, Eloi Schmauch2, Jeffrey Stern1, Karen Khalil1, Jacqueline Kim1, Ian Jaffe1, Marc Lorber4, David Ayares3, Nader Moazami1, Robert A Montgomery1.
1Surgery, NYU Langone Transplant Institute, New York, NY, United States; 2Computer Science and Artificial Intelligence Laboratory, Broad Institute of Harvard and MIT, Cambridge, MA, United States; 3Revivicor Inc, , Blacksburg, VA, United States; 4United Therapeutics, Silver Spring, MD, United States
Background: Advances in xenotransplantation in living and decedent humans using pig xenografts in the last 3 years has shown significnt promise for future compassionate-use and first-in-human trials. Major obstacles remain including incomplete knowledge of the genetic incompatibilities between pig donors and human recipients, which may lead to harmful immune responses against the xenograft and/or physiological dysfunction. In 2022 in NYU, two heart from 10-gene edited pigs were transplanted into two brain-dead human decedents, primarily to evaluate hyper-acute antibody mediated rejection and sustained function of the heart xenograft over a 3 day period.
Methods: We performed multi-omics profiling to assess the dynamic interactions between these two transplanted pig heart-xenografts and the two human decedents. We generated bulk transcriptomic, lipidomic, proteomic and metabolomics, and single-cell RNAsequencing datasets, across blood samples every 6 hrs, as well as histological and transcriptomic tissue profiling to assess global and specific biological changes that correlate with immune related outcomes and xenograft function.
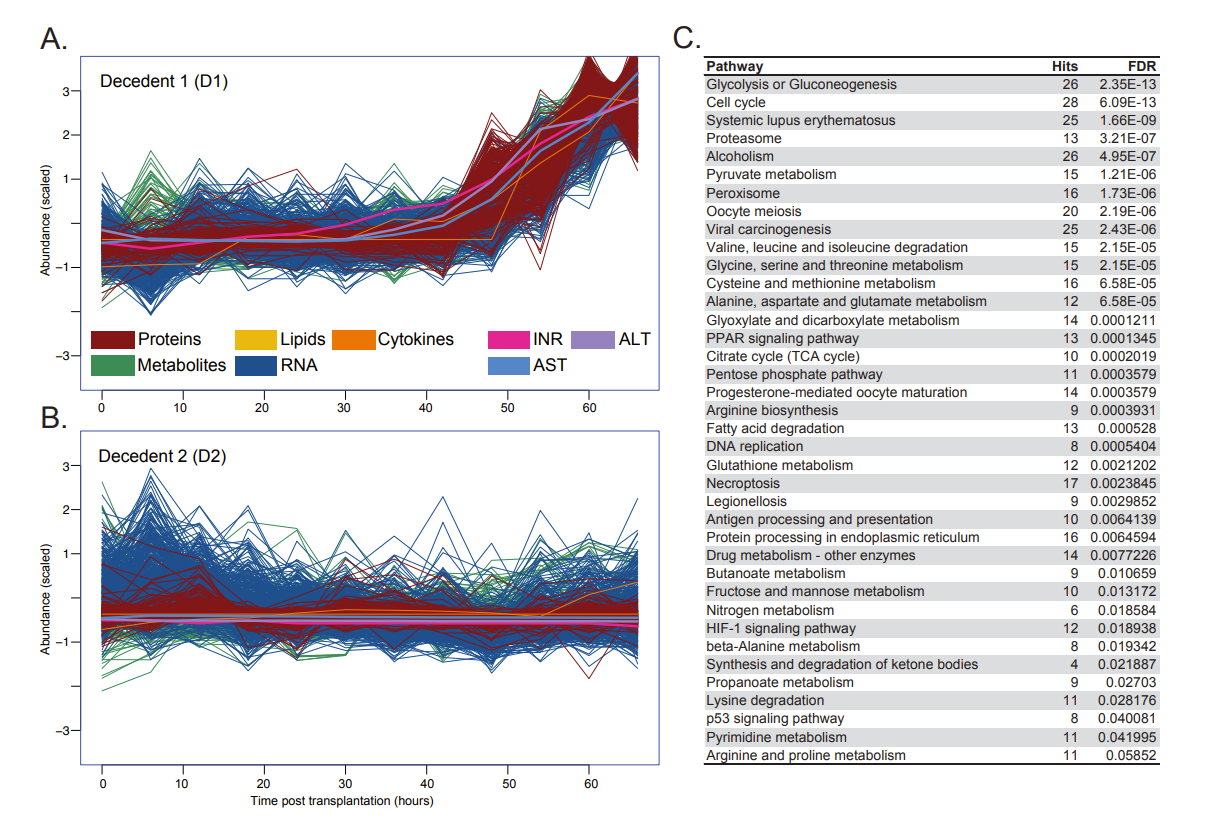
Results: In human decedent 1 we observed specific and significant early immune-activation changes in PBMCs (using scRNAseq) and xenograft tissue (snRNA-seq and spatial transcriptomics) leading to significant downstream T cell and NK cell activity, which collectively represented >20% of all cells in the final Day 3 timepoints. Longitudinal multi-omics integrative analyses from RNAseq, lipidomics, proteomics and metabolomics in blood, and RNAseq in tissue indicated ischemia reperfusion injury (IRI) in decedent 1, which was complicated by inadequate immunosuppression of T cells. This signature is consistent with previous pig to non-human primate perioperative cardiac xenograft dysfunction (PCXD) transcriptomic signatures. This was accompanied by general vascular remodeling signals shared between both decedents. Figure 1 illustrate integrative multiomic analyses of RNA, lipid, protein and metabolite datasets. We also observed significant cellular metabolism and liver damage pathway changes in the latter stage of Day 2 in decedent 1 that correlated with profound organ-wide physiological dysfunction. Decedent 2 had normal xenograft functioning with more moderate changes across the multi-omics profiling datasets.
Conclusions: Single-cell and multi-omics approaches revealed fundamental insights into early molecular and immune responses indicative of IRI and PCXD in human decedent models receiving genetically modified pig heart xenografts, that were not evident in the initial clinical findings.
[1] Big Data, Xenotransplantation
